01:00
Day Two:
Cleaning
~30 min
Overview
Questions
- How basic data import issues can be assessed?
Lesson Objectives
To be able to
- Mange presence/absence/multiple-empty header lines in source data.
- Select best data variable/column names for working with code
- Handle missing data (including empty rows and columns).
Cleaning imported data
Standard data issues
Often, when data where collected in real life in Excel, they do not have an optimal shape for importing them into programs.
You can have:
- leading header’s groups
- empty columns and rows
- multi-line headers
- space and special characters in names
- missing information
- repeated-data reported at first occurrence only

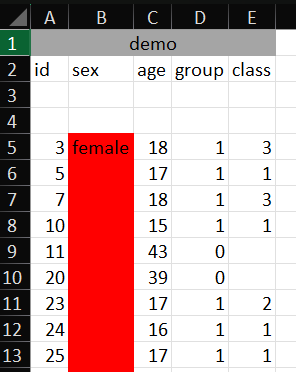
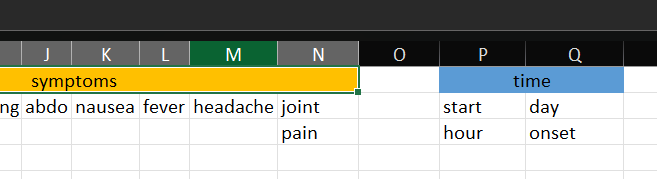
Your turn
- Connect to our pad (https://bit.ly/ubep-rws-pad-ed3) Ex. 13
- List all other issue you have ever encounter in importing/cleaning data (or you can imagine could happen, if this is the first time you import data on software for data analyses)
Headers - skip-them-all
We can completely skip the headers, and manually assign col names
Tip
rio::import useful options:
-
skip = n: ignore first n lines -
header = FALSE= doesn’t use (first-line-after-skipped) data to create headers.
Headers - manual defined headers
We can completely skip the headers, and manually assign col names
Tip
rio::import useful options:
-
skip = n: ignore first n lines -
header = FALSE= doesn’t use (first-line-after-skipped) data to create headers.
[1] "k_1" "d_2" "e_3" "l_4" "r_5" "g_6" "h_7" "y_8"
[9] "r_9" "c_10" "a_11" "o_12" "u_13" "l_14" "k_15" "z_16"
[17] "a_17" "j_18" "v_19" "p_20" "v_21" "v_22" "t_23" "v_24"
[25] "t_25" "j_26" "n_27" "m_28" "q_29" "m_30" "u_31" "z_32"
[33] "h_33" "l_34" "l_35" "i_36" "z_37" "p_38" "g_39" "c_40"
[41] "x_41" "e_42" "n_43"Headers - set manual headers
We can completely skip the headers, and manually assign col names
Tip
rio::import useful options:
-
skip = n: ignore first n lines -
header = FALSE= doesn’t use (first-line-after-skipped) data to create headers.
Headers - {unheadr}
The unheadr package purpose is exactly: “…functions to work with messy data, often derived from spreadsheets…”, in particular, its ?unheadr::mash_colnames makes many header rows into column names.
Important
For unheadr to work, the best option is to rio::import all the data without considering any structure, so that with header = FALSE activated. If so, we can keep n_name_rows = n with n exactly equal to the number of rows that compose the header.
Note
- All the spreadsheet content is imported as data, and all columns are parsed as
characters. - Actual column names are meaningless now, so we will set
keep_names = FALSE.
Headers - {unheadr}
The unheadr package purpose is exactly: “…functions to work with messy data, often derived from spreadsheets…”, in particular, its ?unheadr::mash_colnames makes many header rows into column names.
Important
If we have a (single!) topmost leading rows of grouping column names, and we like to maintain them (otherwise skip = 1), we will activate the option sliding_headers = TRUE. Setting the n_name_rows = n accordingly.
Note
- The group column names are repeated forward to all the empty columns up to the next non empty one.
What’s next
We need to include all the other three header rows in the colnames, merging them with the corresponding group name.
Headers - {unheadr}
The unheadr package purpose is exactly: “…functions to work with messy data, often derived from spreadsheets…”, in particular, its ?unheadr::mash_colnames makes many header rows into column names.
Important
Including in n_name_rows = n all the rows composing the header(s) (topmost grouping one eventually included!), unheadr will merge them all using underscores (i.e., _) to separate words.
Note
- Considering all the rows composing the header is enough to complete the import.
This way, even if multiple column have the same name on distinct groups, we keep distinct column names in our imported dataset!
What’s next
Now that we have merged all the rows reporting column names components, it is extremely useful to guarantee that naming are syntactically correct and with a consistent convention across the dataset.
Variable names
janitor can also convert colnames consistently in a specific convention.1
What’s next
Now colnames are fine, but still some empty column and rows in the dataset, we would like to remove to have a clean structure.
Empty rows and columns
We can use janitor package to remove empty columns and rows.
What’s next
We have finished working on meta-data now. We can start to look at the data them self. First, demo_sex column as implicit repeated values we would like to make explicit.
Fill default content
library(rio)
library(here)
library(unheadr)
library(janitor)
library(tidyverse)
options(rio.import.class = "tibble")
here(
"data-raw",
"Copenhagen_raw.xlsx"
) |>
import(header = FALSE) |>
mash_colnames(
keep_names = FALSE,
n_name_rows = 4,
sliding_headers = TRUE
) |>
clean_names() |>
remove_empty(c("rows", "cols")) |>
fill(demo_sex)What’s next
We are quite to the end, we only need to make consistent convention for missing information. They are both reported as empty (already parsed as NA, correctly) and with ??. We need to tell rio::import that also them are missing information
Missing data
library(rio)
library(here)
library(unheadr)
library(janitor)
library(tidyverse)
options(rio.import.class = "tibble")
here(
"data-raw",
"Copenhagen_raw.xlsx"
) |>
import(
header = FALSE,
na = c("", "??")
) |>
mash_colnames(
keep_names = FALSE,
n_name_rows = 4,
sliding_headers = TRUE
) |>
clean_names() |>
remove_empty(c("rows", "cols")) |>
fill(demo_sex)Your turn (main: A; bk1: B; bk2: C)
Your turn
Connect to the Day-2 project in RStudio cloud (https://bit.ly/ubep-rws-rstudio)
- Open the script
08-clean.Rand follow the instruction step by step.
25:00
Important
Most often you will not have to manage multi-row header nor implicit information. Depending on your data, missing information can be coded not with simple empty cells, so that you would need to set na = c() explicitly. On the other hand, the pattern
library(rio)
library(here)
library(janitor)
db <- here(<path>) |>
import() |>
clean_names() |>
remove_empty(c("rows", "cols"))will be quite standard anytime!
My turn
YOU: Connect to our pad (https://bit.ly/ubep-rws-pad-ed3) and write there questions & doubts (and if I am too slow or too fast)
ME: Connect to the Day-3 project in RStudio cloud (https://bit.ly/ubep-rws-rstudio): script 10-cleaning.R
Homework
Posit’s RStudio Cloud Workspace
Instructions
- Go to: https://bit.ly/ubep-rws-rstudio
Your turn
- Project: day-2
- Instructions:
- Go to: https://bit.ly/ubep-rws-website
- The text is the Day-2 assessment under the tab “Summative Assessments”.
- (on RStudio Cloud)
homework/day_two-summative.html
- Script to complete:
homework/solution.R
Acknowledgment
To create the current lesson, we explored, used, and adapted content from the following resources:
The slides are made using Posit’s Quarto open-source scientific and technical publishing system powered in R by Yihui Xie’s Kintr.
Additional resources
License
This work by Corrado Lanera, Ileana Baldi, and Dario Gregori is licensed under CC BY 4.0
References
Break
10:00

UBEP’s R training for supervisors