15:00
Day Two:
Data structures
~30 min
Overview
Questions
- What kind of objects have R?
- How can I manage them?
- How can I control the flow of code execution?
- How can I define custom functions?
Lesson Objectives
To be able to do/use
- Define
forloops andifconditional executions - Create functions
Data Structures
(atomic) vectors [side]
Atomic vectors are homogeneous/flat objects, i.e. all the objects composing the sequence must be of the same type, and cannot contain other (nested) sequences.
Four main types (?typeof) of atomic vectors:
| Character | ?is.character |
(other) vectors - Factors [side]
Other structure in R are based on atomic vectors, i.e. are of one of the base types but have more structure (similar structures are called ?class)
Factors are discrete (i.e. based on ?integers) variables with labels.
| Factors | ?is.factor |
[1] male female female
Levels: female male otherTip
You can investigate the internal structure of any R objects using ?str.
(other) vectors - Dates / Date-times [side]
Other structure in R are based on atomic vectors, i.e. are of one of the base types but have more structure (similar structures are called ?class)
Dates are counts (based on ?doubles) of days since 1970-01-01.1
| Dates | ?as.Date |
Date-Time are counts (based on ?doubles) of seconds since 1970-01-01.
list (vectors) [side]
List vectors are heterogeneous/nestable objects, i.e. objects composing the sequence can be of distinct types, and can contain other (nested) sequences.
| List | ?list |
$age
[1] 70 85 69
$height
[1] 1.50 1.72 1.81
$at_risk
[1] TRUE FALSE TRUE
$gender
[1] male female female
Levels: female male other(other) lists - data frames (and tibble) [side]
Other structure in R are based on list vectors, i.e. are heterogeneous sequence of objects.
data frames are ordered list of equally sized homogeneous named vectors. I.e. the are used for tabular data:
- ordered list of columns of information, with headers (
?names) - in a column there is one type of information (homogeneous)
- all columns have the same
?length, i.e. number of rows (?nrow)
| Data frames | ?data.frame |
Tip
During the course we will see, explain and use tibbles (from the package {tibble}): a modern, enhanced, better displayed, and with stricter and more consistent structure than standard data frames.
vectors as trains [side]
Important
R works on vectors of two types only:
- Atomic (homogeneous / flat)
- List (heterogeneous / nested)
Think of objects in R (any objects in R!) as a train (either atomic or list) made of wagons:
- a train (i.e., a vector) is sequence of wagons (i.e., objects, homogeneous or heterogeneous, possibly other trains)
- wagons have content (i.e., the data they contain)
- wagons can have labels (i.e., names)
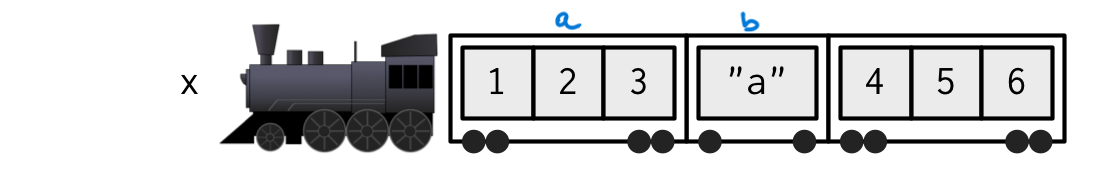
x <- list(a = 1:3, b = "a", 4:6) (image adapted from Advanced-R)
Subsetting - subset [side]
Important
You can refer to subsetting objects (i.e., a train) as performing two operations mainly:
- create another objects (i.e., another train) with a subset of its elements (i.e., wagons)
- extract the content of a (single) object (i.e., the content of a wagon)
You can select more than one object/wagon when subsetting, but a single one only when extracting!
Three ways to identify elements (i.e., wagons):
Subsetting data - coordinates [optional]
Important
A data frame (and tibbles) can be see as a “matrix” (or a table).
Data frames values can be subsetted using “[rows, column]” notation
| multiple selection | [ |
Tip
Use the additional argument drop = FALSE to maintain the data frame structure. Using tibbles we will consistently get always a tibble when subsetting with coordinates!
Subsetting data - extract [side]
Important
You can refer to subsetting objects (i.e., a train) as performing two operations mainly:
- create another objects (i.e., another train) with a subset of its elements (i.e., wagons)
- extract the content of a (single) object (i.e., the content of a wagon)
You can select more than one object/wagon when subsetting, but a single one only when extracting!
Two ways to identify a (single!) element (i.e., a wagon):
- with its position.
- with its name, if it has a name.
Your turn (main: B; bk1: C; bk2: A)
Your turn
Connect to our pad(https://bit.ly/ubep-rws-pad-ed3)
Connect to the Day-2 project in RStudio cloud (https://bit.ly/ubep-rws-rstudio)
Before to evaluate it, in the pad, under the section
2.2. Ex7, write (in a new line) what is your expected result (including an error).Before to evaluate it, in the pad, under the section
2.2. Ex8, write (in a new line) what is your expected result (including an error).Then, open the script
04-atomic-vectors.Rand follow the instruction step by step.Then, open the script
05-subsetting.Rand follow the instruction step by step.
Important
Coercion rule from specific to general:logical > integer > double > character
Subset operation can be performed in sequence on the same object directly.
Crucial to know if you are working on a subset of an object or its content.
My turn
YOU: Connect to our pad(https://bit.ly/ubep-rws-pad-ed3) and write there questions & doubts (and if I am too slow or too fast)
ME: Connect to the Day-2 project in RStudio cloud
- (https://bit.ly/ubep-rws-rstudio): script
02-vectors.R - (https://bit.ly/ubep-rws-rstudio): script
03-matrices_dataframes.R
Control flow [optional]
Tip
You don’t need to test if a logical is TRUE or FALSE, they are already TRUE or FALSE!
Functions [optional]
sum_one <- function(x) {
1 x + 1
}
sum_one(x = 3)- 1
- A function always returns its last evaluated objects.
[1] 4Your turn [optional]
Your turn
Connect to our pad(https://bit.ly/ubep-rws-pad-ed3)
Connect to the Day-2 project in RStudio cloud (https://bit.ly/ubep-rws-rstudio)
- Before to evaluate it, in the pad, under the section
2.2. Ex9, write (in a new line) what is your expected result from the following computation:
- Then, open the script
06-cond-and-funs.Rand follow the instruction step by step.
20:00
Important
?ifreturns the last computed value, if any, orNULLotherwise (which may happen if there is noelse).?forreturnsNULL
My turn [optional]
YOU: Connect to our pad(https://bit.ly/ubep-rws-pad-ed3) and write there questions & doubts (and if I am too slow or too fast)
ME: Connect to the Day-2 project in RStudio cloud (https://bit.ly/ubep-rws-rstudio): script 04-cond_loop.R
Break
10:00
Acknowledgment
To create the current lesson we explored, use, and adapt contents from the following resources:
- Carpentries’ Programming with R course material.
- Carpentries’ R for Reproducible Scientific Analysis course material.
- Hadley Wickham’s Advanced R - (2e)
- Hadley Wickham’s R for Data Science (2e)
The slides are made using Posit’s Quarto open-source scientific and technical publishing system powered in R by Yihui Xie’s kintr.
Additionl Resources
- The Tidyverse
- RStudio cloud tutorials
License
This work by Corrado Lanera, Ileana Baldi, and Dario Gregori is licensed under CC BY 4.0

UBEP’s R training for supervisors