Day Two:
Filter and Select
Datasets
30 min approx
Overview
Questions
- What are Tidy data, why are they useful(, and how to transform untidy data to tidy one)?
- How to select some variables/columns only?
- How to filter rows/cases that match certain conditions?
Lesson Objectives
To be able to
- (Use
pivot_*,separate,unitefunction from the tidyr package in the Tidyverse to reshape data into tidy one.) - Select/filter columns/rows of tibbles (i.e., data frames).
Data shape
Tidy data
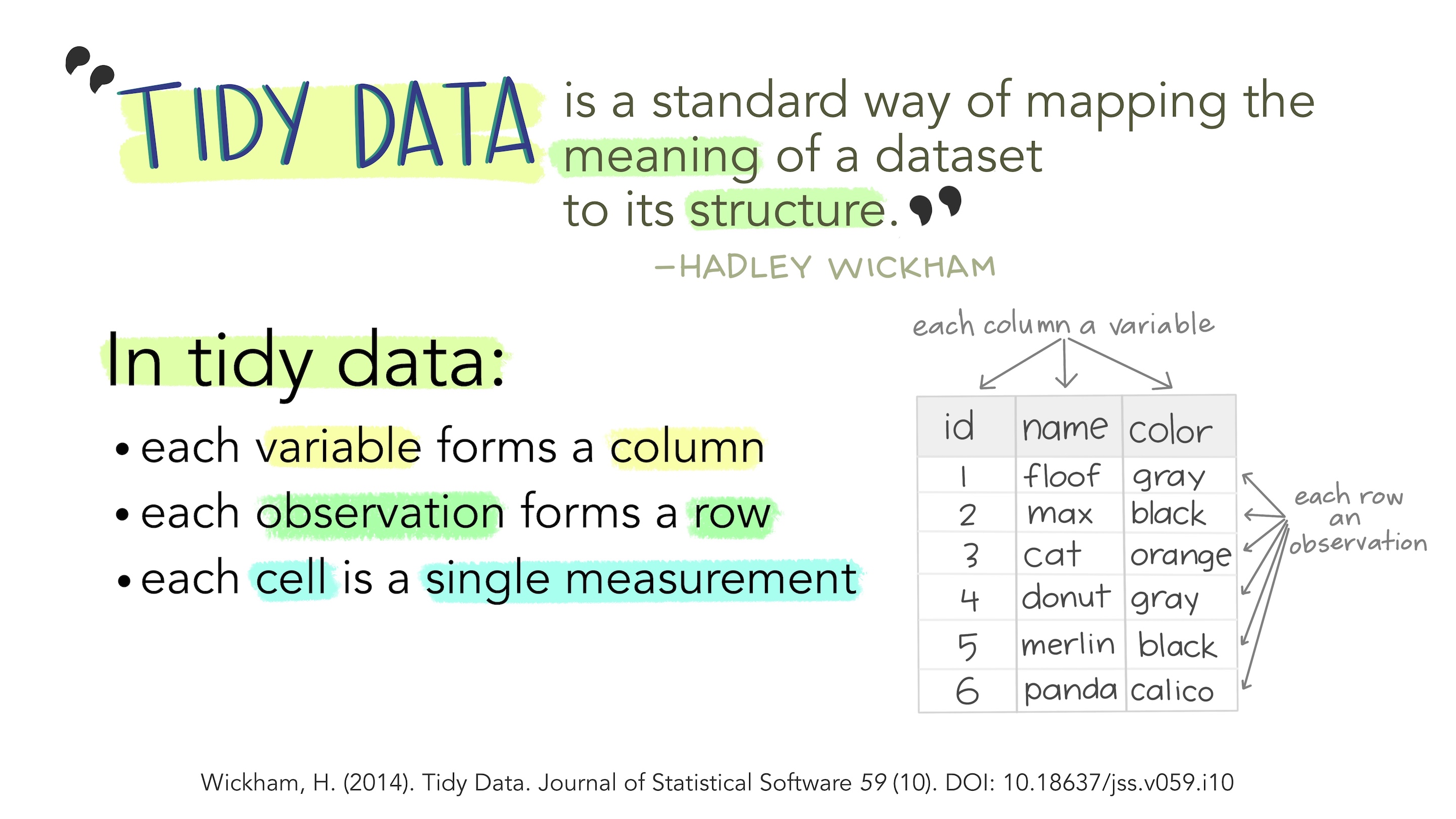
Illustrations from the Openscapes blog Tidy Data for reproducibility, efficiency, and collaboration by Julia Lowndes and Allison Horst
Untidy data
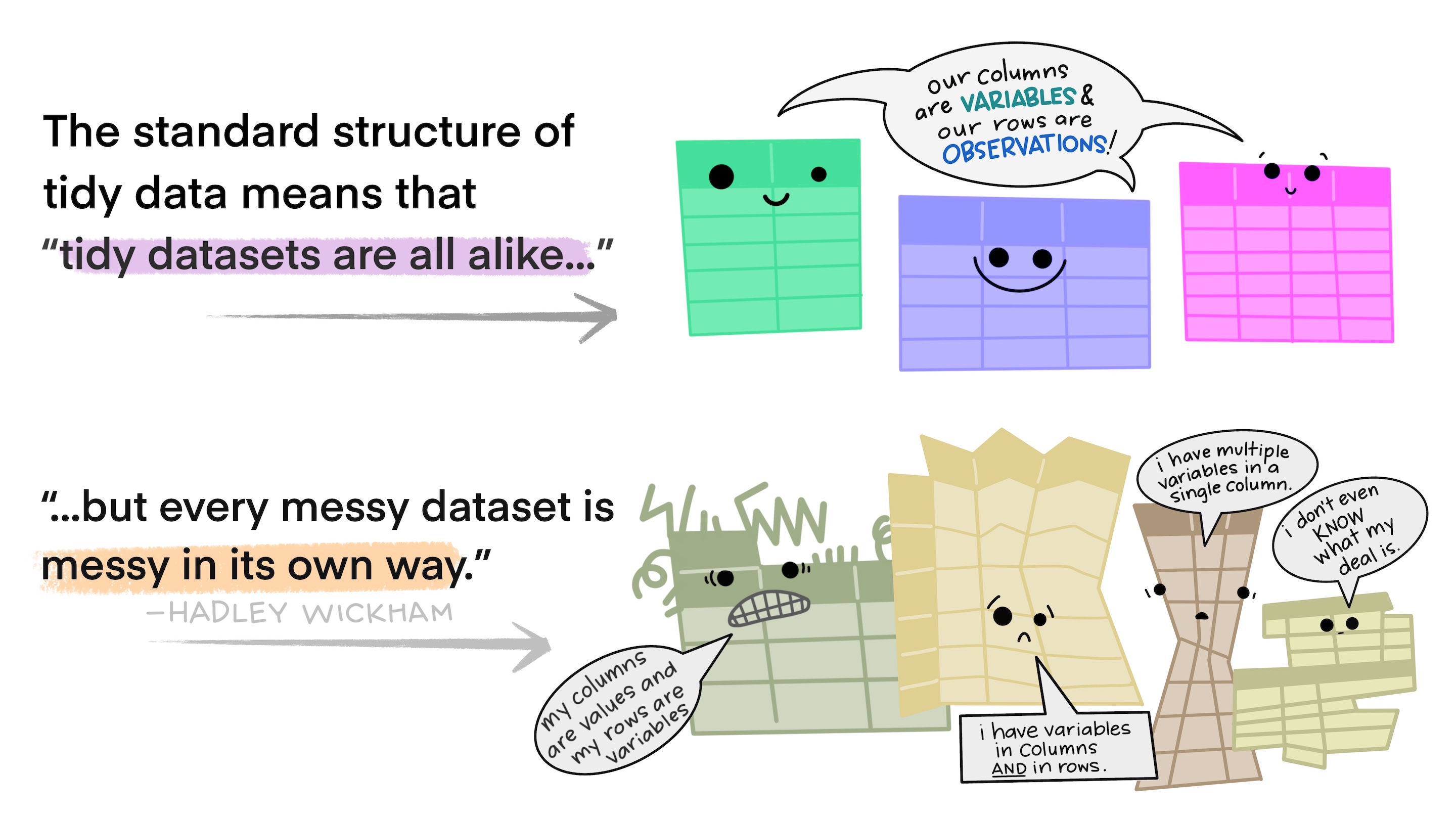
Illustrations from the Openscapes blog Tidy Data for reproducibility, efficiency, and collaboration by Julia Lowndes and Allison Horst
Why tidy data
Illustrations from the Openscapes blog Tidy Data for reproducibility, efficiency, and collaboration by Julia Lowndes and Allison Horst
Tidy rules
There are three interrelated rules that make a dataset tidy:
- Each variable is a column; each column is a variable.
- Each observation is a row; each row is an observation.
- Each value is a cell; each cell is a single value.

Why untidy data
- Data is often organized to facilitate some goal other than analysis. For example, it’s common for data to be structured to make data entry, not analysis, easy.
Example: tidyverse::billboard dataset.1
Warning
- information in column:
-
wk1-wk76should be a single variable: the week. - cell values of
wk1-wk76should be a single variable: the rank.
-
Start Tidying - tidyr::pivot_longer [Optional]
- Data is often organized to facilitate some goal other than analysis. For example, it’s common for data to be structured to make data entry, not analysis, easy.
Important
-
tidyr::pivot_longerconvert your data in “longer” format -
cols: select which variable should be pivoting -
names_to: define the column hosting thecolscolnames -
values_to: define the column hosting thecolsvalues
Warning
Many possibly uninformative missing information!
Start Tidying - tidyr::pivot_longer [Optional]
- Data is often organized to facilitate some goal other than analysis. For example, it’s common for data to be structured to make data entry, not analysis, easy.
Important
-
tidyr::pivot_longerconvert your data in “longer” format -
cols: select which variable should be pivoting -
names_to: define the column hosting thecolscolnames -
values_to: define the column hosting thecolsvalues -
values_drop_na: decide if rows with missing information in values should be removed
Selectors 1
var1:var10: variables lying between var1 on the left and var10 on the right.starts_with("a"): names that start with “a”.ends_with("z"): names that end with “z”.contains("b"): names that contain “b”.matches("x.y"): names that match regular expressionx.y. 2num_range(x, 1:4): names following the pattern,x1,x2, …,x4.all_of(vars)/any_of(vars): names stored in the character vector vars.all_of(vars)will error if the variables aren’t present;any_of(var)will match just the variables that exist.
everything(): all variables.last_col(): furthest column on the right.where(is.numeric): all variables where is.numeric() returns TRUE.
Tip
!selection: only variables that don’t match selection.selection1 & selection2: only variables included in both selection1 and selection2.selection1 | selection2: all variables that match either selection1 or selection2
Multiple variable in colnames [Optional]
Multiple variable in colnames [Optional]
Tip
In case of multiple variable in each colname, you can pivoting them maintaining the underling structure. This way you can separate them in a further second step using tidyr::separate.
Multiple variable in colnames [Optional]
tidyr::pivot_wider [Optional]
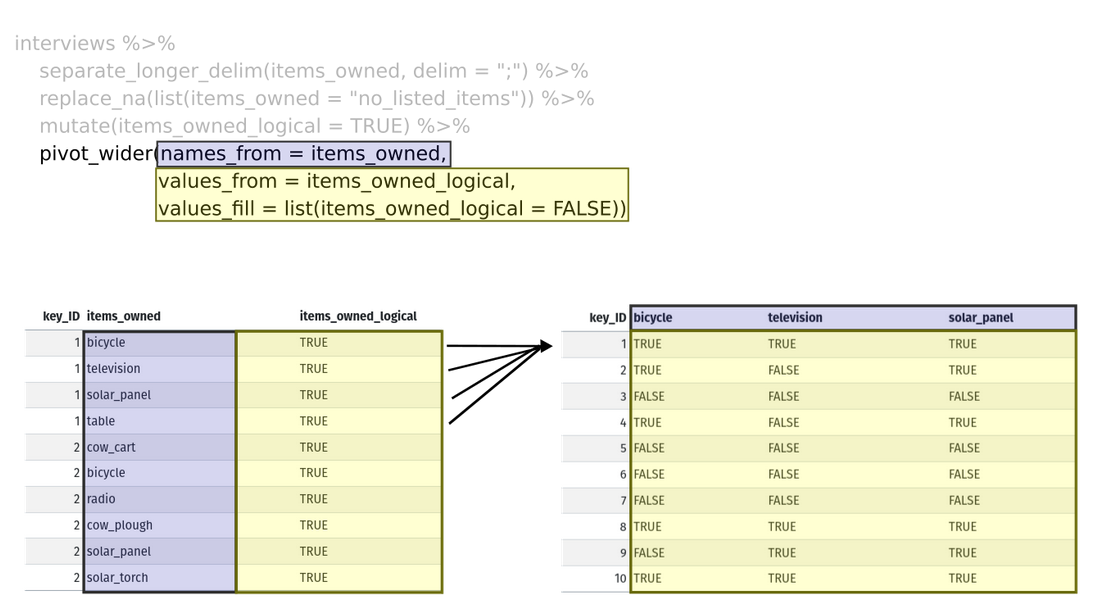
Image from Data Carpentry’s R for Social Scientists
Reverse pivot - tidyr::pivot_wider
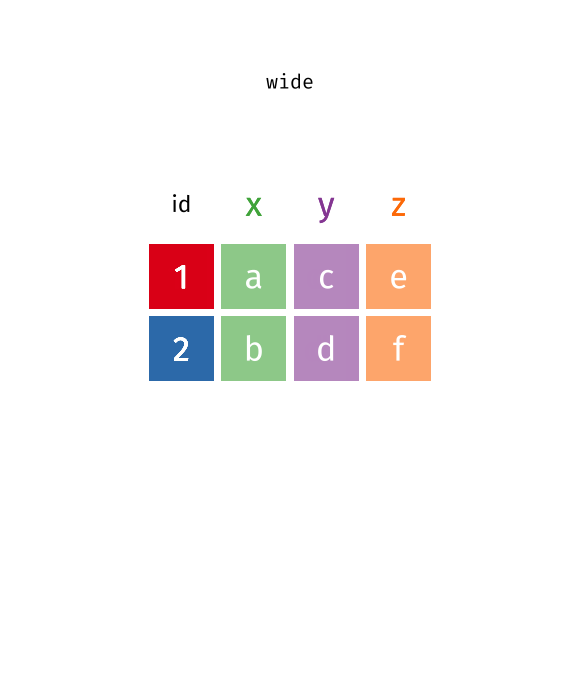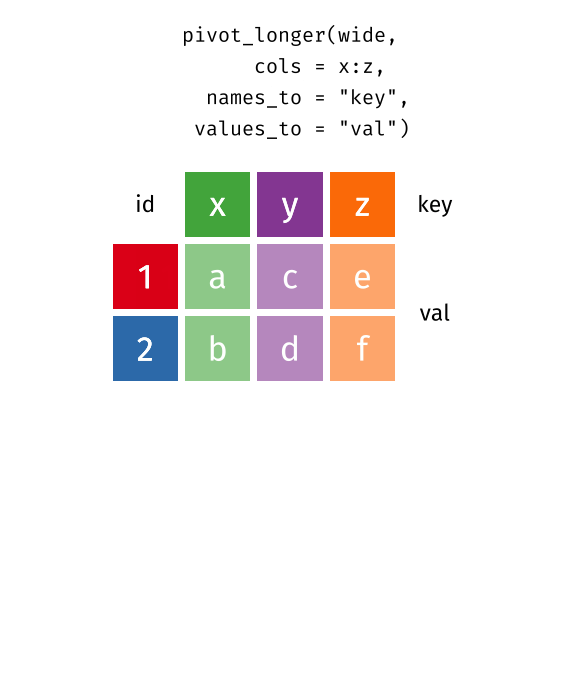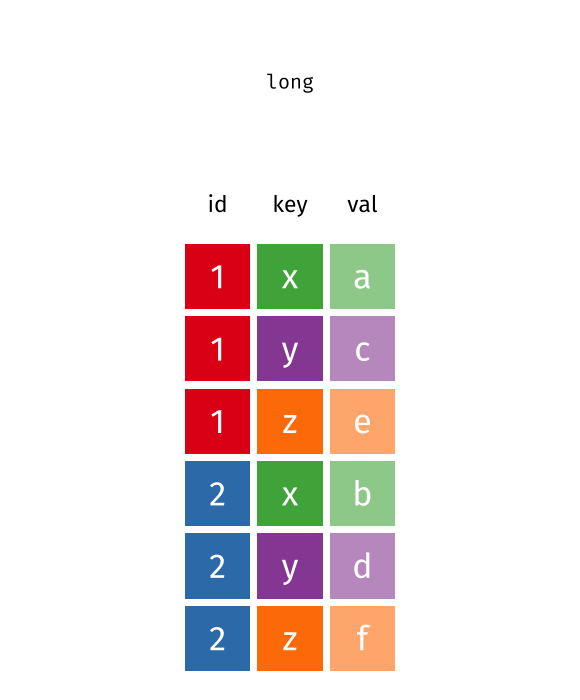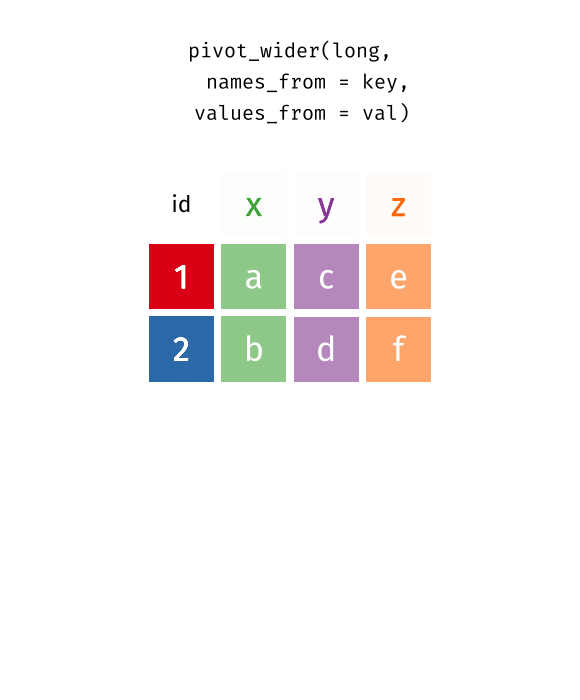
Reverse pivot - example [Optional]
[1] TRUEYour turn [optional]
Your turn
Connect to our pad (https://bit.ly/ubep-rws-pad-ed3) Ex 15-16
Connect to the Day-3 project in RStudio cloud (https://bit.ly/ubep-rws-rstudio)
- Answer in the pad, with an “x” next to the correct answers.
- What are the main option for
pivot_longer? - What are the main option for
pivot_wider?
names_fromnames_to
values_fromvalues_to
- Open the scripts
09-pivot_longer.Rand10-pivot_wider.R, and follow the instruction.
25:00
Important
To transform a table to a longer one, you need to put some of its columns
names_toa new column, and their correspondingvalues_toanother one! Possibly allowingvalues_drop_na.To transform a table to a wider one, you need to take new column
names_froman existing column, and their correspondingvalues_fromthe associated one! Possibly with created missingvalues_filled.
My turn [optional]
YOU: Connect to our pad (https://bit.ly/ubep-rws-pad-ed3) and write there questions & doubts (and if I am too slow or too fast)
ME: Connect to the Day-3 project in RStudio cloud (https://bit.ly/ubep-rws-rstudio): script 11-pivoting.R
Data management
dplyr - intro
Common structure:
- The first argument is always a data frame
- The subsequent arguments typically describe which columns to operate on, using the variable names (without quotes).
- The output is always a new data frame.
Tip
All verbs in Tidyverse are designed to do one thing mainly, and to it well! So, to solve complex problem we will often combine multiple verbs, and we use the pipe (|>) as we are already familiar!
Rows - dplyr::filter [side-by-side]
Important
dplyr::filter allows you to keep rows based on the values of the columns.
Rows - conditions [side-by-side]
We can use any kind of condition inside dplyr::filter; e.g.,
And
Rows - conditions [side-by-side]
We can use any kind of condition inside dplyr::filter; e.g.,
Or
Rows - conditions [side-by-side]
We can use any kind of condition inside dplyr::filter; e.g.,
In
Rows - conditions [side-by-side]
We can use any kind of condition inside dplyr::filter; e.g.,
Not equal
Rows - multiple conditions [side-by-side]
We can also combine together multiple condition of arbitrary complexity at once
Tip
It could be difficult to remind the priority order of logical operators. Using parentheses to group each conditions is a safe way to not be wrong!
Columns - dplyr::select [side-by-side]
For analyses, you do not need to remove columns from your dataset, but it could be extremely useful to see more clearly only the data you need to see time to time.1
You can select the column to keep using the dplyr::select() verb providing:
The variables you like to keep
Columns - dplyr::select [side-by-side]
For analyses, you do not need to remove columns from your dataset, but it could be extremely useful to see more clearly only the data you need to see time to time.1
You can select the column to keep using the dplyr::select() verb providing:
A range of variables you like to keep
Columns - dplyr::select [side-by-side]
For analyses, you do not need to remove columns from your dataset, but it could be extremely useful to see more clearly only the data you need to see time to time.1
You can select the column to keep using the dplyr::select() verb providing:
Excludig the selection (!)
Columns - dplyr::select [side-by-side]
For analyses, you do not need to remove columns from your dataset, but it could be extremely useful to see more clearly only the data you need to see time to time.1
You can select the column to keep using the dplyr::select() verb providing:
Matching a condition - where
Selectors 1 [side-by-side]
var1:var10: variables lying between var1 on the left and var10 on the right.starts_with("a"): names that start with “a”.ends_with("z"): names that end with “z”.contains("b"): names that contain “b”.matches("x.y"): names that match regular expressionx.y. 2num_range(x, 1:4): names following the pattern,x1,x2, …,x4.all_of(vars)/any_of(vars): names stored in the character vector vars.all_of(vars)will error if the variables aren’t present;any_of(var)will match just the variables that exist.
everything(): all variables.last_col(): furthest column on the right.where(is.numeric): all variables where is.numeric() returns TRUE.
Tip
!selection: only variables that don’t match selection.selection1 & selection2: only variables included in both selection1 and selection2.selection1 | selection2: all variables that match either selection1 or selection2
Your turn (main: A; bk1: B; bk2: C)
Your turn
Connect to our pad(https://bit.ly/ubep-rws-pad-ed3)
Connect to the Day-3 project in RStudio cloud (https://bit.ly/ubep-rws-rstudio)
Answer in the pad, under the section
3.2. Ex17, and3.2. Ex18.Then, open the script
11-filter.Rand12-select.R, and follow the instruction.
20:00
Important
- First argument of
dplyr::filteranddplyr::selectis always a data frame - Returned object of
dplyr::filteranddplyr::selectis always a data frame - Original data frame won’t be modified by
dplyr::filternordplyr::select, never!
Important
- you can put arbitrary complex conditions returning logical vectors of the same length of the number of rows of the data frame, involving any column of the data frame in use also.
-
all_of(vec)is for strict selection. If any of the variables in the charactervecis missing, an error is thrown. -
any_of(vec)doesn’t check for missing variables. It is especially useful with negative selections, when you would like to make sure a variable is removed.
My turn
YOU: Connect to our pad (https://bit.ly/ubep-rws-pad-ed3) and write there questions & doubts (and if I am too slow or too fast)
ME: Connect to the Day-3 project in RStudio cloud (https://bit.ly/ubep-rws-rstudio): script 12-filter-and-select.R
Acknowledgment
To create the current lesson, we explored, used, and adapted content from the following resources:
The slides are made using Posit’s Quarto open-source scientific and technical publishing system powered in R by Yihui Xie’s Kintr.
Additional resources
License
This work by Corrado Lanera, Ileana Baldi, and Dario Gregori is licensed under CC BY 4.0
References

UBEP’s R training for supervisors