[1] "data.frame"
Day One:
Import
~20 min
Overview
Questions
- How data can be imported end exported from/to files on disk?
Lesson Objectives
To be able to
- import and export data from/to files on disk
Import/Export
Data file
For doing real data analyses, we need to interact with the external world with data. Reading and writing them.
Mainly, we will work with the so-called rectangular data. I.e., information that can be organized in a tables:
- all columns (AKA variables) have the same number of rows.
- all rows (AKA observations) have the same number of columns
- each data value correspond to a single row-column pair within the table.

Reminder on data frames overview
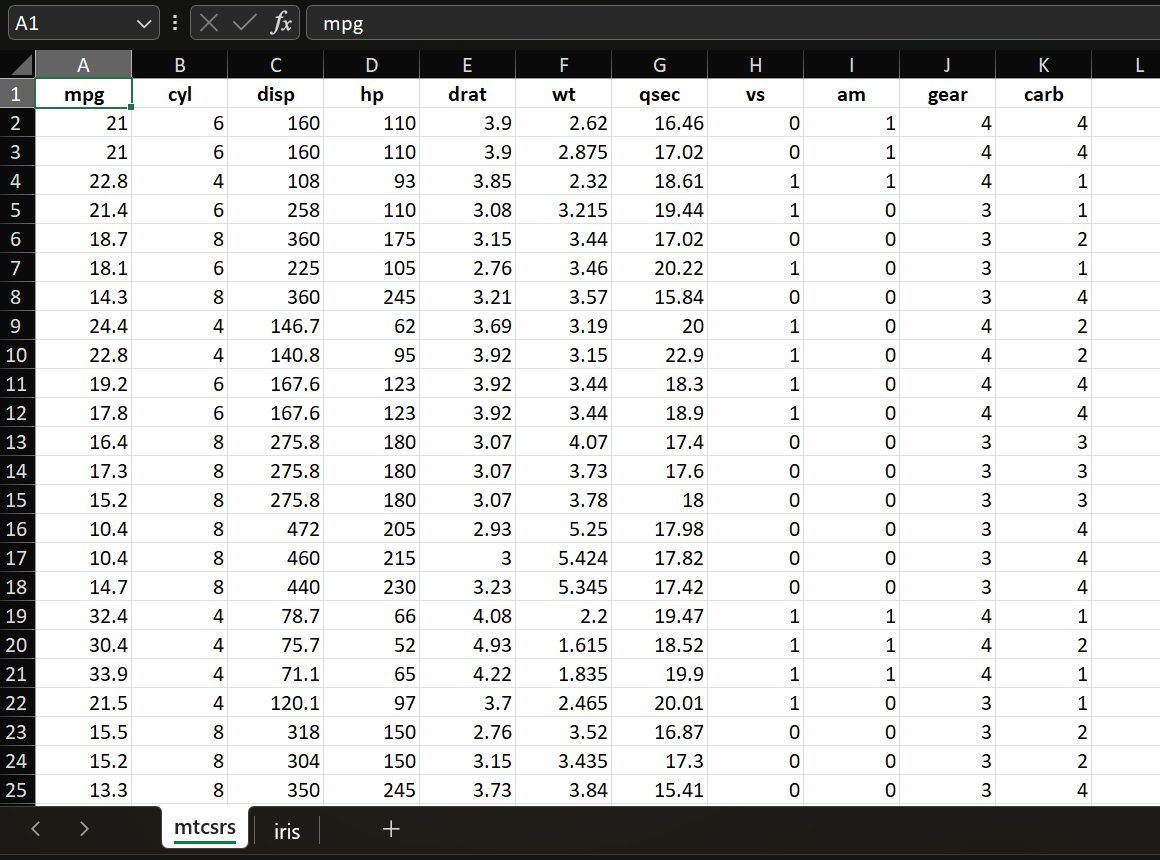
R tabular data structure is the data.frame
Warning
Row names are not data, but an attribute of the data frame, so as_tibble will remove them.
mpg cyl disp hp drat wt qsec vs am
Mazda RX4 21.0 6 160.0 110 3.90 2.620 16.46 0 1
Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1
Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1
Hornet 4 Drive 21.4 6 258.0 110 3.08 3.215 19.44 1 0
Hornet Sportabout 18.7 8 360.0 175 3.15 3.440 17.02 0 0
Valiant 18.1 6 225.0 105 2.76 3.460 20.22 1 0
Duster 360 14.3 8 360.0 245 3.21 3.570 15.84 0 0
Merc 240D 24.4 4 146.7 62 3.69 3.190 20.00 1 0
Merc 230 22.8 4 140.8 95 3.92 3.150 22.90 1 0
Merc 280 19.2 6 167.6 123 3.92 3.440 18.30 1 0
Merc 280C 17.8 6 167.6 123 3.92 3.440 18.90 1 0
Merc 450SE 16.4 8 275.8 180 3.07 4.070 17.40 0 0
Merc 450SL 17.3 8 275.8 180 3.07 3.730 17.60 0 0
Merc 450SLC 15.2 8 275.8 180 3.07 3.780 18.00 0 0
Cadillac Fleetwood 10.4 8 472.0 205 2.93 5.250 17.98 0 0
Lincoln Continental 10.4 8 460.0 215 3.00 5.424 17.82 0 0
Chrysler Imperial 14.7 8 440.0 230 3.23 5.345 17.42 0 0
Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1
Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1
Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1
Toyota Corona 21.5 4 120.1 97 3.70 2.465 20.01 1 0
Dodge Challenger 15.5 8 318.0 150 2.76 3.520 16.87 0 0
AMC Javelin 15.2 8 304.0 150 3.15 3.435 17.30 0 0
Camaro Z28 13.3 8 350.0 245 3.73 3.840 15.41 0 0
Pontiac Firebird 19.2 8 400.0 175 3.08 3.845 17.05 0 0
Fiat X1-9 27.3 4 79.0 66 4.08 1.935 18.90 1 1
Porsche 914-2 26.0 4 120.3 91 4.43 2.140 16.70 0 1
Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.90 1 1
Ford Pantera L 15.8 8 351.0 264 4.22 3.170 14.50 0 1
Ferrari Dino 19.7 6 145.0 175 3.62 2.770 15.50 0 1
Maserati Bora 15.0 8 301.0 335 3.54 3.570 14.60 0 1
Volvo 142E 21.4 4 121.0 109 4.11 2.780 18.60 1 1
gear carb
Mazda RX4 4 4
Mazda RX4 Wag 4 4
Datsun 710 4 1
Hornet 4 Drive 3 1
Hornet Sportabout 3 2
Valiant 3 1
Duster 360 3 4
Merc 240D 4 2
Merc 230 4 2
Merc 280 4 4
Merc 280C 4 4
Merc 450SE 3 3
Merc 450SL 3 3
Merc 450SLC 3 3
Cadillac Fleetwood 3 4
Lincoln Continental 3 4
Chrysler Imperial 3 4
Fiat 128 4 1
Honda Civic 4 2
Toyota Corolla 4 1
Toyota Corona 3 1
Dodge Challenger 3 2
AMC Javelin 3 2
Camaro Z28 3 4
Pontiac Firebird 3 2
Fiat X1-9 4 1
Porsche 914-2 5 2
Lotus Europa 5 2
Ford Pantera L 5 4
Ferrari Dino 5 6
Maserati Bora 5 8
Volvo 142E 4 2In the tidyverse, we will use a modern version of the data frame called tibble (class: tbl_df)
# A tibble: 32 × 11
mpg cyl disp hp drat wt qsec vs am gear
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 21 6 160 110 3.9 2.62 16.5 0 1 4
2 21 6 160 110 3.9 2.88 17.0 0 1 4
3 22.8 4 108 93 3.85 2.32 18.6 1 1 4
4 21.4 6 258 110 3.08 3.22 19.4 1 0 3
5 18.7 8 360 175 3.15 3.44 17.0 0 0 3
6 18.1 6 225 105 2.76 3.46 20.2 1 0 3
7 14.3 8 360 245 3.21 3.57 15.8 0 0 3
8 24.4 4 147. 62 3.69 3.19 20 1 0 4
9 22.8 4 141. 95 3.92 3.15 22.9 1 0 4
10 19.2 6 168. 123 3.92 3.44 18.3 1 0 4
# ℹ 22 more rows
# ℹ 1 more variable: carb <dbl>Reminder on data frames [optional]
R tabular data structure is the data.frame
Warning
Row names are not data, but an attribute of the data frame, so as_tibble will remove them.
To keep row names, use rownames = "<name>".
mpg cyl disp hp drat wt qsec vs am
Mazda RX4 21.0 6 160.0 110 3.90 2.620 16.46 0 1
Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1
Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1
Hornet 4 Drive 21.4 6 258.0 110 3.08 3.215 19.44 1 0
Hornet Sportabout 18.7 8 360.0 175 3.15 3.440 17.02 0 0
Valiant 18.1 6 225.0 105 2.76 3.460 20.22 1 0
Duster 360 14.3 8 360.0 245 3.21 3.570 15.84 0 0
Merc 240D 24.4 4 146.7 62 3.69 3.190 20.00 1 0
Merc 230 22.8 4 140.8 95 3.92 3.150 22.90 1 0
Merc 280 19.2 6 167.6 123 3.92 3.440 18.30 1 0
Merc 280C 17.8 6 167.6 123 3.92 3.440 18.90 1 0
Merc 450SE 16.4 8 275.8 180 3.07 4.070 17.40 0 0
Merc 450SL 17.3 8 275.8 180 3.07 3.730 17.60 0 0
Merc 450SLC 15.2 8 275.8 180 3.07 3.780 18.00 0 0
Cadillac Fleetwood 10.4 8 472.0 205 2.93 5.250 17.98 0 0
Lincoln Continental 10.4 8 460.0 215 3.00 5.424 17.82 0 0
Chrysler Imperial 14.7 8 440.0 230 3.23 5.345 17.42 0 0
Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1
Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1
Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1
Toyota Corona 21.5 4 120.1 97 3.70 2.465 20.01 1 0
Dodge Challenger 15.5 8 318.0 150 2.76 3.520 16.87 0 0
AMC Javelin 15.2 8 304.0 150 3.15 3.435 17.30 0 0
Camaro Z28 13.3 8 350.0 245 3.73 3.840 15.41 0 0
Pontiac Firebird 19.2 8 400.0 175 3.08 3.845 17.05 0 0
Fiat X1-9 27.3 4 79.0 66 4.08 1.935 18.90 1 1
Porsche 914-2 26.0 4 120.3 91 4.43 2.140 16.70 0 1
Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.90 1 1
Ford Pantera L 15.8 8 351.0 264 4.22 3.170 14.50 0 1
Ferrari Dino 19.7 6 145.0 175 3.62 2.770 15.50 0 1
Maserati Bora 15.0 8 301.0 335 3.54 3.570 14.60 0 1
Volvo 142E 21.4 4 121.0 109 4.11 2.780 18.60 1 1
gear carb
Mazda RX4 4 4
Mazda RX4 Wag 4 4
Datsun 710 4 1
Hornet 4 Drive 3 1
Hornet Sportabout 3 2
Valiant 3 1
Duster 360 3 4
Merc 240D 4 2
Merc 230 4 2
Merc 280 4 4
Merc 280C 4 4
Merc 450SE 3 3
Merc 450SL 3 3
Merc 450SLC 3 3
Cadillac Fleetwood 3 4
Lincoln Continental 3 4
Chrysler Imperial 3 4
Fiat 128 4 1
Honda Civic 4 2
Toyota Corolla 4 1
Toyota Corona 3 1
Dodge Challenger 3 2
AMC Javelin 3 2
Camaro Z28 3 4
Pontiac Firebird 3 2
Fiat X1-9 4 1
Porsche 914-2 5 2
Lotus Europa 5 2
Ford Pantera L 5 4
Ferrari Dino 5 6
Maserati Bora 5 8
Volvo 142E 4 2In the tidyverse, we will use a modern version of the data frame called tibble (class: tbl_df)
# A tibble: 32 × 12
model mpg cyl disp hp drat wt qsec vs am
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Mazda R… 21 6 160 110 3.9 2.62 16.5 0 1
2 Mazda R… 21 6 160 110 3.9 2.88 17.0 0 1
3 Datsun … 22.8 4 108 93 3.85 2.32 18.6 1 1
4 Hornet … 21.4 6 258 110 3.08 3.22 19.4 1 0
5 Hornet … 18.7 8 360 175 3.15 3.44 17.0 0 0
6 Valiant 18.1 6 225 105 2.76 3.46 20.2 1 0
7 Duster … 14.3 8 360 245 3.21 3.57 15.8 0 0
8 Merc 24… 24.4 4 147. 62 3.69 3.19 20 1 0
9 Merc 230 22.8 4 141. 95 3.92 3.15 22.9 1 0
10 Merc 280 19.2 6 168. 123 3.92 3.44 18.3 1 0
# ℹ 22 more rows
# ℹ 2 more variables: gear <dbl>, carb <dbl>Plain Vs. non-plain text data
Tabular data can be (quite always) saved as a plain text, readable by anyone, on every computer.
Most common plain-text tabular data file format is “CSV”, i.e., Comma-Separated Value.
Extension for those files is .csv (e.g., data.csv)
Warning
In EU, we use a comma to separate decimal digits instead of a dot. For this reason, it would be ambiguous to use comma to separate field in CSV, which must use quotes in those cases. For this reason, there is an alternative version of CSV, called CSV2 (using the same file extension .csv!!) which uses a semicolon to separate fields and can use comma for numbers.
On the other hand, it could be difficult to collect data in plain text, and often they are collected by other software, e.g., Excel, in non-plain text format.
Extension for those files is .xlsx (e.g., data.xlsx)
Base R [optional]
- read CSV with
?read.csv
id sex age group class diarrhoea bloody vomiting abdo
1 1 male 18 student 2 TRUE FALSE FALSE TRUE
2 3 female 18 student 3 NA NA NA NA
3 5 female 17 student 1 NA NA NA TRUE
4 6 male 17 student 2 NA NA NA NA
5 7 female 18 student 3 TRUE FALSE FALSE TRUE
6 8 male 18 student 2 TRUE FALSE FALSE TRUE
nausea fever headache jointpain starthour meal tuna tunaD
1 FALSE NA FALSE FALSE 9 TRUE TRUE 2
2 NA NA NA NA NA TRUE FALSE 0
3 TRUE NA TRUE NA NA TRUE NA NA
4 NA NA NA NA NA TRUE FALSE 0
5 TRUE FALSE TRUE FALSE 15 TRUE TRUE 2
6 FALSE FALSE FALSE FALSE 15 TRUE TRUE 2
shrimps shrimpsD green greenD veal vealD pasta pastaD rocket
1 TRUE 2 FALSE 0 TRUE 2 TRUE 3 TRUE
2 FALSE 0 FALSE 0 TRUE 1 TRUE 3 TRUE
3 NA NA NA NA TRUE 0 TRUE 1 NA
4 FALSE 0 FALSE 0 TRUE 0 FALSE 0 FALSE
5 TRUE 2 TRUE 2 TRUE 2 TRUE 2 TRUE
6 TRUE 2 TRUE 2 TRUE 2 TRUE 2 TRUE
rocketD sauce sauceD bread breadD champagne champagneD beer
1 1 TRUE 2 TRUE 2 TRUE 1 TRUE
2 3 TRUE 3 TRUE 3 TRUE 1 FALSE
3 NA NA NA TRUE 1 FALSE 0 FALSE
4 0 FALSE 0 FALSE 0 TRUE 3 TRUE
5 2 TRUE 2 TRUE 2 TRUE 1 TRUE
6 2 TRUE 2 TRUE 2 TRUE 1 TRUE
beerD redwine redwineD whitewine whitewineD
1 3 FALSE 0 FALSE 0
2 0 TRUE 3 FALSE 0
3 0 FALSE 0 FALSE 0
4 3 TRUE 3 TRUE 3
5 2 FALSE 0 TRUE 3
6 3 FALSE 0 TRUE 2
dayonset onset_datetime meal_datetime
1 2006-11-12T00:00:00Z 2006-11-12T09:00:00Z 2006-11-11T18:00:00Z
2 2006-11-11T18:00:00Z
3 2006-11-11T18:00:00Z
4 2006-11-11T18:00:00Z
5 2006-11-12T00:00:00Z 2006-11-12T15:00:00Z 2006-11-11T18:00:00Z
6 2006-11-13T00:00:00Z 2006-11-13T15:00:00Z 2006-11-11T18:00:00Z
gastrosymptoms ate_anything case incubation
1 TRUE TRUE TRUE 15
2 FALSE TRUE FALSE NA
3 FALSE TRUE FALSE NA
4 FALSE TRUE FALSE NA
5 TRUE TRUE TRUE 21
6 TRUE TRUE TRUE 45 Student.ID Full.Name favourite.food
1 1 Sunil Huffmann Strawberry yoghurt
2 2 Barclay Lynn French fries
3 3 Jayendra Lyne N/A
4 4 Leon Rossini Anchovies
5 5 Chidiegwu Dunkel Pizza
6 6 Güvenç Attila Ice cream
mealPlan AGE
1 Lunch only 4
2 Lunch only 5
3 Breakfast and lunch 7
4 Lunch only
5 Breakfast and lunch five
6 Lunch only 6Tidyverse [optional]
- read CSV with
?read_csv
# A tibble: 384 × 46
id sex age group class diarrhoea bloody vomiting abdo
<dbl> <chr> <dbl> <chr> <dbl> <lgl> <lgl> <lgl> <lgl>
1 1 male 18 stud… 2 TRUE FALSE FALSE TRUE
2 3 female 18 stud… 3 NA NA NA NA
3 5 female 17 stud… 1 NA NA NA TRUE
4 6 male 17 stud… 2 NA NA NA NA
5 7 female 18 stud… 3 TRUE FALSE FALSE TRUE
6 8 male 18 stud… 2 TRUE FALSE FALSE TRUE
7 9 male 61 teac… NA NA NA NA NA
8 10 female 15 stud… 1 FALSE FALSE FALSE FALSE
9 11 female 43 teac… NA TRUE NA NA NA
10 12 male 16 stud… 1 NA NA NA NA
# ℹ 374 more rows
# ℹ 37 more variables: nausea <lgl>, fever <lgl>,
# headache <lgl>, jointpain <lgl>, starthour <dbl>,
# meal <lgl>, tuna <lgl>, tunaD <dbl>, shrimps <lgl>,
# shrimpsD <dbl>, green <lgl>, greenD <dbl>, veal <lgl>,
# vealD <dbl>, pasta <lgl>, pastaD <dbl>, rocket <lgl>,
# rocketD <dbl>, sauce <lgl>, sauceD <dbl>, bread <lgl>, …# A tibble: 6 × 5
`Student ID` `Full Name` favourite.food mealPlan AGE
<dbl> <chr> <chr> <chr> <chr>
1 1 Sunil Huffmann Strawberry yoghurt Lunch o… 4
2 2 Barclay Lynn French fries Lunch o… 5
3 3 Jayendra Lyne N/A Breakfa… 7
4 4 Leon Rossini Anchovies Lunch o… <NA>
5 5 Chidiegwu Dunkel Pizza Breakfa… five
6 6 Güvenç Attila Ice cream Lunch o… 6 Non plain-text data [optional]
# A tibble: 384 × 46
id sex age group class diarrhoea bloody vomiting abdo
<dbl> <chr> <dbl> <chr> <chr> <lgl> <lgl> <lgl> <lgl>
1 1 male 18 stud… 2 TRUE FALSE FALSE TRUE
2 3 female 18 stud… 3 NA NA NA NA
3 5 female 17 stud… 1 NA NA NA TRUE
4 6 male 17 stud… 2 NA NA NA NA
5 7 female 18 stud… 3 TRUE FALSE FALSE TRUE
6 8 male 18 stud… 2 TRUE FALSE FALSE TRUE
7 9 male 61 teac… <NA> NA NA NA NA
8 10 female 15 stud… 1 FALSE FALSE FALSE FALSE
9 11 female 43 teac… <NA> TRUE NA NA NA
10 12 male 16 stud… 1 NA NA NA NA
# ℹ 374 more rows
# ℹ 37 more variables: nausea <lgl>, fever <lgl>,
# headache <lgl>, jointpain <lgl>, starthour <dbl>,
# meal <lgl>, tuna <lgl>, tunaD <dbl>, shrimps <lgl>,
# shrimpsD <dbl>, green <lgl>, greenD <dbl>, veal <lgl>,
# vealD <dbl>, pasta <lgl>, pastaD <dbl>, rocket <lgl>,
# rocketD <dbl>, sauce <lgl>, sauceD <dbl>, bread <lgl>, …# A tibble: 32 × 6
YEAR Y W R L K
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1948 1.21 0.243 0.145 1.41 0.612
2 1949 1.35 0.260 0.218 1.38 0.559
3 1950 1.57 0.278 0.316 1.39 0.573
4 1951 1.95 0.297 0.394 1.55 0.564
5 1952 2.27 0.310 0.356 1.80 0.574
6 1953 2.73 0.322 0.359 1.93 0.711
7 1954 3.03 0.335 0.403 1.96 0.776
8 1955 3.56 0.350 0.396 2.12 0.827
9 1956 3.98 0.361 0.382 2.43 0.800
10 1957 4.42 0.379 0.305 2.71 0.921
# ℹ 22 more rows{rio}
From one side, it could be better to use tidyverse functions (i.e., readr ones, which is part of the Tidyverse) to read tabular data into R because of a more consistent naming and arguments.
But, we still need haven to read SAS, SPSS, STATA, and other types of data, and we need readxl to read Excel files. Moreover, we still need to recognize and link the file type with the function used to read it.
Tip
We can use the rio package to read them all!
?rio::importprovides a painless data import experience by automatically choosing the appropriate import/read function based on file extension (or a specified format argument)?rio::exportprovides the same painless file recognition for data export/write functionality
{rio} - read [side-by-side]
id sex age group class diarrhoea bloody vomiting abdo
1 1 male 18 student 2 TRUE FALSE FALSE TRUE
2 3 female 18 student 3 NA NA NA NA
3 5 female 17 student 1 NA NA NA TRUE
4 6 male 17 student 2 NA NA NA NA
5 7 female 18 student 3 TRUE FALSE FALSE TRUE
6 8 male 18 student 2 TRUE FALSE FALSE TRUE
nausea fever headache jointpain starthour meal tuna tunaD
1 FALSE NA FALSE FALSE 9 TRUE TRUE 2
2 NA NA NA NA NA TRUE FALSE 0
3 TRUE NA TRUE NA NA TRUE NA NA
4 NA NA NA NA NA TRUE FALSE 0
5 TRUE FALSE TRUE FALSE 15 TRUE TRUE 2
6 FALSE FALSE FALSE FALSE 15 TRUE TRUE 2
shrimps shrimpsD green greenD veal vealD pasta pastaD rocket
1 TRUE 2 FALSE 0 TRUE 2 TRUE 3 TRUE
2 FALSE 0 FALSE 0 TRUE 1 TRUE 3 TRUE
3 NA NA NA NA TRUE 0 TRUE 1 NA
4 FALSE 0 FALSE 0 TRUE 0 FALSE 0 FALSE
5 TRUE 2 TRUE 2 TRUE 2 TRUE 2 TRUE
6 TRUE 2 TRUE 2 TRUE 2 TRUE 2 TRUE
rocketD sauce sauceD bread breadD champagne champagneD beer
1 1 TRUE 2 TRUE 2 TRUE 1 TRUE
2 3 TRUE 3 TRUE 3 TRUE 1 FALSE
3 NA NA NA TRUE 1 FALSE 0 FALSE
4 0 FALSE 0 FALSE 0 TRUE 3 TRUE
5 2 TRUE 2 TRUE 2 TRUE 1 TRUE
6 2 TRUE 2 TRUE 2 TRUE 1 TRUE
beerD redwine redwineD whitewine whitewineD dayonset
1 3 FALSE 0 FALSE 0 2006-11-12
2 0 TRUE 3 FALSE 0 <NA>
3 0 FALSE 0 FALSE 0 <NA>
4 3 TRUE 3 TRUE 3 <NA>
5 2 FALSE 0 TRUE 3 2006-11-12
6 3 FALSE 0 TRUE 2 2006-11-13
onset_datetime meal_datetime gastrosymptoms
1 2006-11-12 09:00:00 2006-11-11 18:00:00 TRUE
2 <NA> 2006-11-11 18:00:00 FALSE
3 <NA> 2006-11-11 18:00:00 FALSE
4 <NA> 2006-11-11 18:00:00 FALSE
5 2006-11-12 15:00:00 2006-11-11 18:00:00 TRUE
6 2006-11-13 15:00:00 2006-11-11 18:00:00 TRUE
ate_anything case incubation
1 TRUE TRUE 15
2 TRUE FALSE NA
3 TRUE FALSE NA
4 TRUE FALSE NA
5 TRUE TRUE 21
6 TRUE TRUE 45# A tibble: 384 × 46
id sex age group class diarrhoea bloody vomiting abdo
<dbl> <chr> <dbl> <chr> <chr> <lgl> <lgl> <lgl> <lgl>
1 1 male 18 stud… 2 TRUE FALSE FALSE TRUE
2 3 female 18 stud… 3 NA NA NA NA
3 5 female 17 stud… 1 NA NA NA TRUE
4 6 male 17 stud… 2 NA NA NA NA
5 7 female 18 stud… 3 TRUE FALSE FALSE TRUE
6 8 male 18 stud… 2 TRUE FALSE FALSE TRUE
7 9 male 61 teac… <NA> NA NA NA NA
8 10 female 15 stud… 1 FALSE FALSE FALSE FALSE
9 11 female 43 teac… <NA> TRUE NA NA NA
10 12 male 16 stud… 1 NA NA NA NA
# ℹ 374 more rows
# ℹ 37 more variables: nausea <lgl>, fever <lgl>,
# headache <lgl>, jointpain <lgl>, starthour <dbl>,
# meal <lgl>, tuna <lgl>, tunaD <dbl>, shrimps <lgl>,
# shrimpsD <dbl>, green <lgl>, greenD <dbl>, veal <lgl>,
# vealD <dbl>, pasta <lgl>, pastaD <dbl>, rocket <lgl>,
# rocketD <dbl>, sauce <lgl>, sauceD <dbl>, bread <lgl>, …# A tibble: 32 × 6
YEAR Y W R L K
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1948 1.21 0.243 0.145 1.41 0.612
2 1949 1.35 0.260 0.218 1.38 0.559
3 1950 1.57 0.278 0.316 1.39 0.573
4 1951 1.95 0.297 0.394 1.55 0.564
5 1952 2.27 0.310 0.356 1.80 0.574
6 1953 2.73 0.322 0.359 1.93 0.711
7 1954 3.03 0.335 0.403 1.96 0.776
8 1955 3.56 0.350 0.396 2.12 0.827
9 1956 3.98 0.361 0.382 2.43 0.800
10 1957 4.42 0.379 0.305 2.71 0.921
# ℹ 22 more rows… and many other
{rio} - write [side-by-side]
# A tibble: 384 × 46
id sex age group class diarrhoea bloody vomiting abdo
<int> <chr> <int> <chr> <int> <lgl> <lgl> <lgl> <lgl>
1 1 male 18 stud… 2 TRUE FALSE FALSE TRUE
2 3 female 18 stud… 3 NA NA NA NA
3 5 female 17 stud… 1 NA NA NA TRUE
4 6 male 17 stud… 2 NA NA NA NA
5 7 female 18 stud… 3 TRUE FALSE FALSE TRUE
6 8 male 18 stud… 2 TRUE FALSE FALSE TRUE
7 9 male 61 teac… NA NA NA NA NA
8 10 female 15 stud… 1 FALSE FALSE FALSE FALSE
9 11 female 43 teac… NA TRUE NA NA NA
10 12 male 16 stud… 1 NA NA NA NA
# ℹ 374 more rows
# ℹ 37 more variables: nausea <lgl>, fever <lgl>,
# headache <lgl>, jointpain <lgl>, starthour <int>,
# meal <lgl>, tuna <lgl>, tunaD <int>, shrimps <lgl>,
# shrimpsD <int>, green <lgl>, greenD <int>, veal <lgl>,
# vealD <int>, pasta <lgl>, pastaD <int>, rocket <lgl>,
# rocketD <int>, sauce <lgl>, sauceD <int>, bread <lgl>, …# A tibble: 384 × 46
id sex age group class diarrhoea bloody vomiting abdo
<dbl> <chr> <dbl> <chr> <dbl> <lgl> <lgl> <lgl> <lgl>
1 1 male 18 stud… 2 TRUE FALSE FALSE TRUE
2 3 female 18 stud… 3 NA NA NA NA
3 5 female 17 stud… 1 NA NA NA TRUE
4 6 male 17 stud… 2 NA NA NA NA
5 7 female 18 stud… 3 TRUE FALSE FALSE TRUE
6 8 male 18 stud… 2 TRUE FALSE FALSE TRUE
7 9 male 61 teac… NA NA NA NA NA
8 10 female 15 stud… 1 FALSE FALSE FALSE FALSE
9 11 female 43 teac… NA TRUE NA NA NA
10 12 male 16 stud… 1 NA NA NA NA
# ℹ 374 more rows
# ℹ 37 more variables: nausea <lgl>, fever <lgl>,
# headache <lgl>, jointpain <lgl>, starthour <dbl>,
# meal <lgl>, tuna <lgl>, tunaD <dbl>, shrimps <lgl>,
# shrimpsD <dbl>, green <lgl>, greenD <dbl>, veal <lgl>,
# vealD <dbl>, pasta <lgl>, pastaD <dbl>, rocket <lgl>,
# rocketD <dbl>, sauce <lgl>, sauceD <dbl>, bread <lgl>, …# A tibble: 384 × 46
id sex age group class diarrhoea bloody vomiting abdo
<dbl> <chr> <dbl> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 male 18 stud… 2 1 0 0 1
2 3 female 18 stud… 3 NA NA NA NA
3 5 female 17 stud… 1 NA NA NA 1
4 6 male 17 stud… 2 NA NA NA NA
5 7 female 18 stud… 3 1 0 0 1
6 8 male 18 stud… 2 1 0 0 1
7 9 male 61 teac… NA NA NA NA NA
8 10 female 15 stud… 1 0 0 0 0
9 11 female 43 teac… NA 1 NA NA NA
10 12 male 16 stud… 1 NA NA NA NA
# ℹ 374 more rows
# ℹ 37 more variables: nausea <dbl>, fever <dbl>,
# headache <dbl>, jointpain <dbl>, starthour <dbl>,
# meal <dbl>, tuna <dbl>, tunaD <dbl>, shrimps <dbl>,
# shrimpsD <dbl>, green <dbl>, greenD <dbl>, veal <dbl>,
# vealD <dbl>, pasta <dbl>, pastaD <dbl>, rocket <dbl>,
# rocketD <dbl>, sauce <dbl>, sauceD <dbl>, bread <dbl>, …# A tibble: 384 × 46
id sex age group class diarrhoea bloody vomiting abdo
<dbl> <chr> <dbl> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 male 18 stud… 2 1 0 0 1
2 3 female 18 stud… 3 NA NA NA NA
3 5 female 17 stud… 1 NA NA NA 1
4 6 male 17 stud… 2 NA NA NA NA
5 7 female 18 stud… 3 1 0 0 1
6 8 male 18 stud… 2 1 0 0 1
7 9 male 61 teac… NA NA NA NA NA
8 10 female 15 stud… 1 0 0 0 0
9 11 female 43 teac… NA 1 NA NA NA
10 12 male 16 stud… 1 NA NA NA NA
# ℹ 374 more rows
# ℹ 37 more variables: nausea <dbl>, fever <dbl>,
# headache <dbl>, jointpain <dbl>, starthour <dbl>,
# meal <dbl>, tuna <dbl>, tunaD <dbl>, shrimps <dbl>,
# shrimpsD <dbl>, green <dbl>, greenD <dbl>, veal <dbl>,
# vealD <dbl>, pasta <dbl>, pastaD <dbl>, rocket <dbl>,
# rocketD <dbl>, sauce <dbl>, sauceD <dbl>, bread <dbl>, …# A tibble: 384 × 46
id sex age group class diarrhoea bloody vomiting abdo
<dbl> <chr> <dbl> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 male 18 stud… 2 1 0 0 1
2 3 female 18 stud… 3 NA NA NA NA
3 5 female 17 stud… 1 NA NA NA 1
4 6 male 17 stud… 2 NA NA NA NA
5 7 female 18 stud… 3 1 0 0 1
6 8 male 18 stud… 2 1 0 0 1
7 9 male 61 teac… NA NA NA NA NA
8 10 female 15 stud… 1 0 0 0 0
9 11 female 43 teac… NA 1 NA NA NA
10 12 male 16 stud… 1 NA NA NA NA
# ℹ 374 more rows
# ℹ 37 more variables: nausea <dbl>, fever <dbl>,
# headache <dbl>, jointpain <dbl>, starthour <dbl>,
# meal <dbl>, tuna <dbl>, tunaD <dbl>, shrimps <dbl>,
# shrimpsD <dbl>, green <dbl>, greenD <dbl>, veal <dbl>,
# vealD <dbl>, pasta <dbl>, pastaD <dbl>, rocket <dbl>,
# rocketD <dbl>, sauce <dbl>, sauceD <dbl>, bread <dbl>, …
{rio} - multiple-sheets Excel [optional]
You can directly save a multi-sheet Excel file writing a list of data frames…
{rio} - multiple-sheets Excel [optional]
… and import multiple sheet data in a (list of) data frame(s)
list of dataframes, from single multi-sheets file
List of 2
$ mtcars: tibble [32 × 11] (S3: tbl_df/tbl/data.frame)
$ iris : tibble [150 × 5] (S3: tbl_df/tbl/data.frame)single dataframe, from multiple ones binded by rows
(from multiple files w/ same colnames)
Your turn (main: A; bk1: B; bk2: C)
Your turn
Connect to our pad(https://bit.ly/ubep-rws-pad-3ed)
Connect to the Day-1 project in RStudio cloud (https://bit.ly/ubep-rws-rstudio)
-
Which function(s) can you use to read excel data from disk?
import_excelreadimport
Then, open the script
08-rio.Rand follow the instruction step by step.
15:00
Important
- you can use
rio::importto read tabular data, it will process the file properly based on its extension and content. - you can use
rio::exportto write tabular data (most of format), simply providing the correct extension.
My turn
YOU: Connect to our pad (https://bit.ly/ubep-rws-pad-ed2) and write there questions & doubts (and if I am too slow or too fast)
ME: Connect to the course-scripts project in RStudio cloud (https://bit.ly/ubep-rws-rstudio): script 09-import.R
Homework
Posit’s RStudio Cloud Workspace
Instructions
Your turn
- Project: day-1
- Instructions (on RStudio Cloud):
- Text:
homework/day_one-summative.html - Script to complete:
homework/solution.R
- Text:
- Solution:
- Go to: https://bit.ly/ubep-rws-website
- The text is the Day-1 assessment under the tab “Summative Assessments”.
Acknowledgment
To create the current lesson, we explored, used, and adapted content from the following resources:
rio package.
The slides are made using Posit’s Quarto open-source scientific and technical publishing system powered in R by Yihui Xie’s Kintr.
License
This work by Corrado Lanera, Ileana Baldi, and Dario Gregori is licensed under CC BY 4.0

UBEP’s R training for supervisors