20:00
Day One:
Infrastructures
~30 min
Overview
Questions
- What is an RStudio project?
- Why using RStudio projects helps?
- How works the here package, and why it is useful in combination with RStudio projects.
- What is a library (and a repository) of R packages, and how many of them we (can) have on a system?
- Why organize a project in standard folders?
- What can be a suitable standard project folder organization?
Lesson Objectives
To be able to
- Activate, restore, a work on an RStudio projects.
- Use the here package to find files and folders within a project.
- Describe the difference from a library and a package, in R.
- Organize a project in folder, in particular understand and be able to use the standard structure of
R/,data-raw/, anddata/, with optional folders foranalyses/, anddev/
R/RStudio projects
Scripts - interface
- You are not required to use the interactive R console alone
- You can save what you type/code for future usage in scripts, i.e., simple text files containing code. If R scripts, their extension is
.R, e.g.,my-first-script.R.
Tip
Strive to create scripts that are working as expected while executed as a whole from top to bottom in a new clean R session.
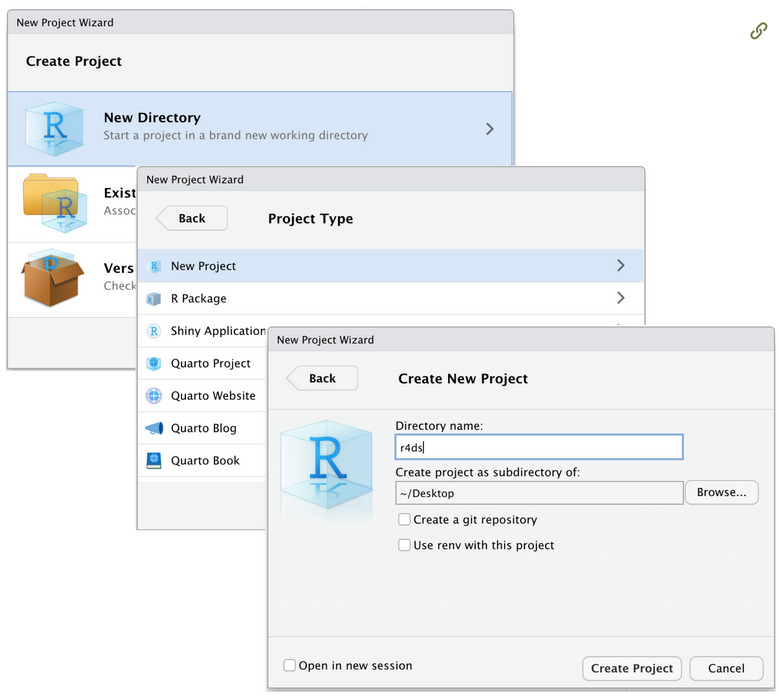
Projects [side-by-side cloud and local]
With RStudio projects, R automatically set the working directory of your session at the project folder. Moreover RStudio automatically save the status of your projects script, including open tabs.
So, with rstudio projects, you can
close and reopen your projects without losing your script (you will lose your R environmental object you have created with your code, but you can always restore it re-running your scripts!).
run multiple R session simultaneously, each one linked to its own working directory, i.e. working effectively on multiple projects.
send/store your projects out from your computer and it will still working
Working directory [side-by-side cloud and local]
Your scripts (with your data) are the source of truth regarding your analyses project!
You/other can and should be able to recreate all your result from your script and data
Where are your analyses?
Every R session is automatically linked to a so called working directory, i.e., a folder in your computer.
Every time you as R to write/save to, read/load from your disk, R will start looking from that folder.
Paths
Absolute
Inside projects you do not need to use (and you should never do that!) use absolute path in reading or writing files and folders.
Absolute paths points to a specific folder in a specific computer, and will never work on other systems, or if you move your project on a different folder of the same PC.
E.g., C:\Users\<usr>\Documents\2023-ecdc-rws\_day-one\slide\
Relative
Inside projects you can (and you should always do that!) use relative path in reading or writing files and folders.
Relative paths point to a path relative to the current working directory, so that they always works on different PCs, or if you move your project folder on other position within your computer.
E.g., _day0one\slide\
On UNIX Machine (linux/mac) path are separated by slashes (e.g., path/to/folder), while on Windows they are separated by back-slashes (e.g., path\to\folder).
However, in R (and many other software), the backslash has special meaning, so that if you need to write a windows-like path in R, you should type them twice every time (e.g., path\\to\\folder).
Tip
On the other hand, R understand and can manage both the standards in all the systems, so that, in R, you can always use the UNIX-like path, even on Windows machines.
The {here} package [side-by-side]
Sometimes the working directory can change with or without our control.1
To use here in your scripts, simply attach it!
-
here()function always uses project-relative paths -
you can compose paths without worrying about the slash/backslash to use!
Your turn (main: C; bk1: A; bk2: B)
Your turn (If you havn’t installed R/RStudio yet locally, do it now with our support)
Connect RStudio cloud (https://bit.ly/ubep-rws-rstudio)
- Create a new project (on the cloud), and pretend you are developing some code for a new analysis.
- Create a new file called
hello.txtand write your name in it. - Create a new script called
read-and-write.Rand write in it the following code
Run the script, what happens? Where is the
hello-me.txtfile?Delete the
hello-me.txtfile, export the project, and unzip it in your computer on the desktop.Double click on the
read-and-write.Rscript directly (RStudio will open it automatically), and run it. What happens? Whare is thehello-me.txtfile? Delete thehello-me.txtfile, and close R/RStudio.Create a folder in your local project named
scripts/, and move theread-and-write.Rscript in it.Double click on the
read-and-write.Rscript now, and run it. What happens? Where is thehello-me.txtfile? Are you able now to run it properly?Change the content of
read-and-write.Rto the following
- Close R/RStudio (locally), double click on the
read-and-write.Rscript, and execute it. What happens? Where is thehello-me.txtfile? Are you able now to run it properly? - Close R/RStudio (locally), move the
read-and-write.Rscript back to the main project folder, and double click on theread-and-write.Rscript, and execute it. What happens? Where is thehello-me.txtfile? Are you able now to run it properly?
My turn
YOU: Connect to our pad(https://bit.ly/ubep-rws-pad-3ed) and write there questions & doubts (and if I am too slow or too fast)
ME: Connect to the Day-1 project in RStudio cloud (https://bit.ly/ubep-rws-rstudio): script 05-packages.R
Best practice
Scripts - shortcuts [optional]
Create new R script with the shortcut
CTRL/CMD + SHIFT + NIn RStudio, you can run/execute/evaluate/send-to-R a complete chunk of code by placing the cursor wherever inside that chunk, and using the shortcut
CTRL/CMD + RETURN.1
Tip
- New script:
CTRL/CMD + SHIFT + N - Run line/chunk of code:
CTRL/CMD + RETURN - Run selected piece/lines/blocks of code: select them and
CTRL/CMD + RETURN - Run (source) the whole script:
CTRL/CMD + SHIFT + S - Restart the R session:
CTRL/CMD + SHIFT + F10
Tip
- Always start your script attaching all used packages, i.e. including all the
library()statements (Easily see which package you would need to install, and what is used to run every piece of code in the script). - Never include (uncommented)
install.packages()statements in a script (especially if you share it! changing other people environment can hamper the systems).
Saving and naming files and folder
Machine readable: avoid spaces, symbols, and special characters.1
Human readable: use file names to describe what’s in the file.
Play well with default ordering: start file names with numbers so that alphabetical sorting puts them in the order they get used.2
Common name
alternative model.R
code for exploratory analysis.r
finalreport.qmd
FinalReport.qmd
fig 1.png
Figure_02.png
model_first_try.R
run-first.r
temp.txtBetter names
01-load-data.R
02-exploratory-analysis.R
03-model-approach-1.R
04-model-approach-2.R
fig-01.png
fig-02.png
report-2022-03-20.qmd
report-2022-04-02.qmd
report-draft-notes.txtTip
Spend time to write/style names and codes that make as fast as possible to understand them. You will spend a lot more time reading, understanding, debugging your code, files, and projects than the amount of time you will pass actually typing them. So, saving time in typing possibly more immediate and faster, but less readable code or less meaningful names is the best way to spend more time overall on projects.
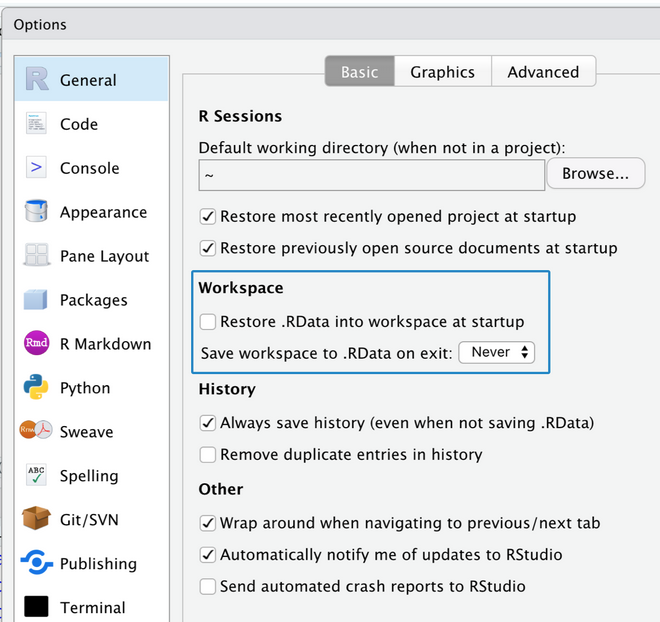
RStudio settings for reproducibility [side-by-side]
Important
From a well designed script and data, all the environmental objects can be recreated
From a well created environmental set of objects, is still quite impossible to reconstruct the code used to create it!
Tip
Do not let R to save/restore your workspace (the default), start every R session with a clean environment!
Folder organization
Now that you:
- have a main project folder to work inside
- can write scripts to store code used for you analyses
- (can write/read files to/from the disk)
- can define robust path relative to the project main folder
You are free to organize and move your scripts and data in well designed folder structures
Tip
Some of the standard R folders are:
-
data-raw/: to store raw data, the original ones they send us -
data/: processed data we will use into our analyses -
R/: a folder to store custom functions source code only (advanced topic)
Suggested folder organization
-
R/: if any, custom functions defined and used within the project. -
data-raw/: to store raw data (i.e., the original ones they send us), and the scripts to preprocess them in preparation for the analyses. -
data/: processed data we will use into our analyses. -
analysis/: analysis scripts. -
output/: report, figure and table produced.
Tip
If you create an R/functions.R script into which you define custom functions, you can make them available during analyses computations including, after the library() calls at the top of the script, the following code:
And next, you can use your custom function within the script.
Tip
source is a base R function that allows to run a whole script, i.e., to execute all the code contained in it.
Break
10:00
Acknowledgment
To create the current lesson, we explored, used, and adapted content from the following resources:
{here}website
The slides are made using Posit’s Quarto open-source scientific and technical publishing system powered in R by Yihui Xie’s Kintr.
Additional resources
License
This work by Corrado Lanera, Ileana Baldi, and Dario Gregori is licensed under CC BY 4.0

UBEP’s R training for supervisors