Day Three:
Transform
Types
30 (+30) min approx
Overview
Questions
- How to handle factors effectively in R/Tidyverse?
- How to handle dates and time in R/Tidyverse?
- How to handle strings in R/Tidyverse?
Lesson Objectives
To be able to
- perform basic factor data management.
- convert textual date/time into date/time R objects
- use simple regular expression and main
str_*functions to manage strings
Mange principal formats
Factors - why
Using strings for categories is not always the best choice. Factors are the best way to represent categories in R.
- sorting issues
- missing/wrong levels issues
- tabulation issues
Factors - how
Define a set of possible values (levels), as a standard character vector.
[1] "Jan" "Feb" "Mar" "Apr" "May" "Jun" "Jul" "Aug" "Sep" "Oct"
[11] "Nov" "Dec"And define a variable as factor, specifying the levels using that.
Base
Tidyverse - (forcats)
Factors - why tidyverse ({forcats})
If we don’t provide explicit levels, the levels are the unique values in the vector, sorted alphabetically in base R, or in the order of appearance in forcats.
Tidyverse - (forcats)
If there are wrong values in the values used to create a factor, they are included as missing (NA) in base R silently, while forcats throws an (informative!) error.
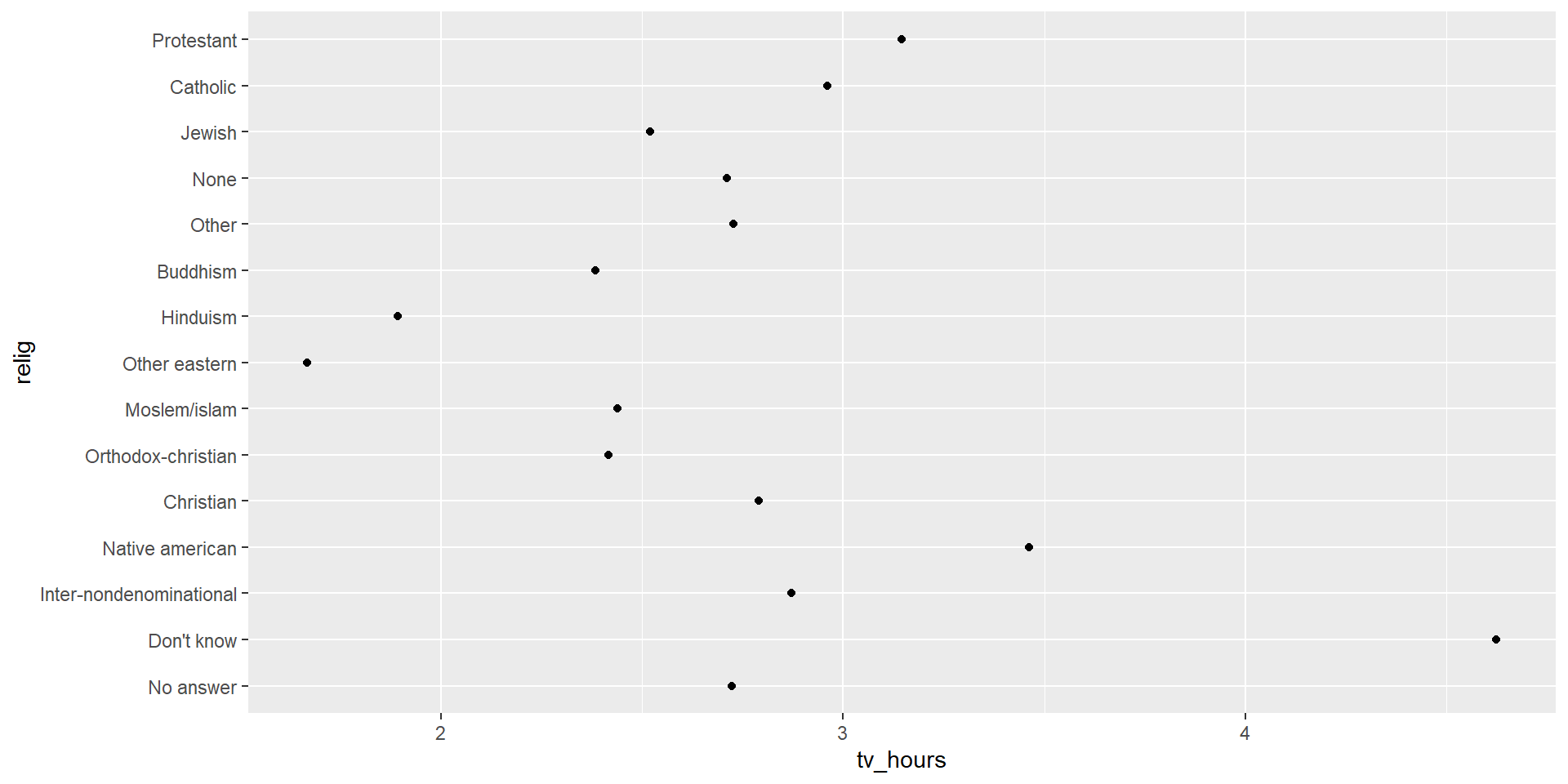
Factors - reorder levels
It could be useful to reordering levels, e.g. when plotting information.
We can use forcats::fct_relevel to reorder levels. Its first argument is the factor to reorder, and the following argument is a numeric vector you want to use to reorder the levels.
Tip
Often, the numerical value you use to reorder a factor is another variable in your dataset!
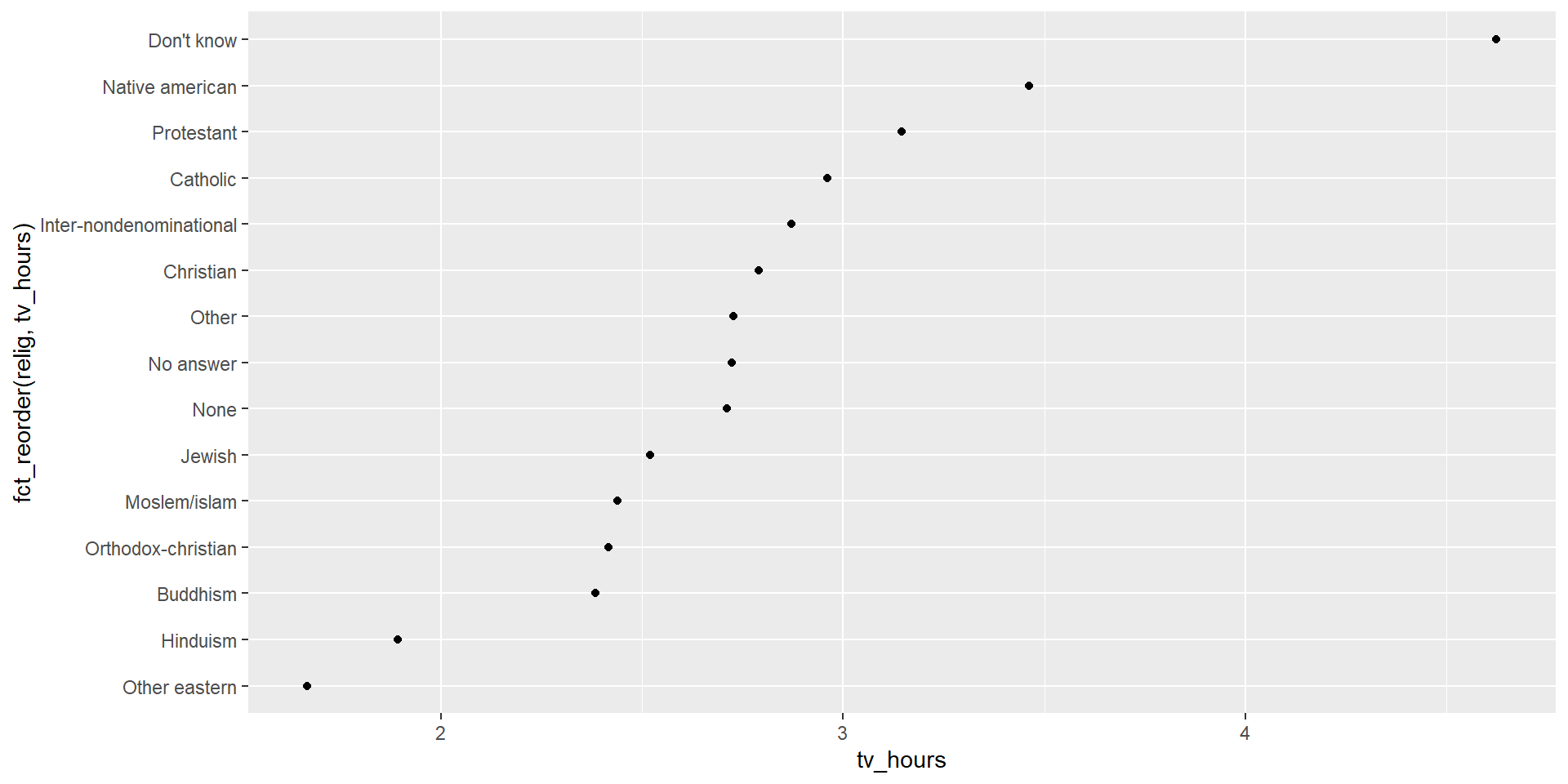
Factors - reorder levels
It could be useful to reordering levels, e.g. when plotting information.
We can use forcats::fct_relevel to reorder levels. Its first argument is the factor to reorder, and the following argument is a numeric vector you want to use to reorder the levels.
Tip
Often, the numerical value you use to reorder a factor is another variable in your dataset!
Factors - reorder levels
It could be useful to reordering levels, e.g. when plotting information.
We can use forcats::fct_relevel to reorder levels. Its first argument is the factor to reorder, and the following argument is a numeric vector you want to use to reorder the levels.
Tip
Often, the numerical value you use to reorder a factor is another variable in your dataset!
Factors - reorder levels
There are also many other useful functions in forcats to reorder levels, e.g., fct_infreq and fct_rev. To see all of them, refer to its website https://forcats.tidyverse.org/.
Factors - modify (AKA recode) levels
We can also modify levels, e.g., to change the wording, or to merge some of them together.
Change the wording
gss_cat |>
mutate(
partyid = fct_recode(partyid,
"Republican, strong" = "Strong republican",
"Republican, weak" = "Not str republican",
"Independent, near rep" = "Ind,near rep",
"Independent, near dem" = "Ind,near dem",
"Democrat, weak" = "Not str democrat",
"Democrat, strong" = "Strong democrat",
"Other" = "No answer",
"Other" = "Don't know",
"Other" = "Other party"
)
) |>
count(partyid)Important
forcats::fct_recode will leave the levels that aren’t explicitly mentioned as is, and will warn you if you accidentally refer to a level that doesn’t exist.
To combine groups, you can assign multiple old levels to the same new level, or… use forcats::fct_collapse!
Factors - modify (AKA recode) levels
We can also modify levels, e.g., to change the wording, or to merge some of them together.
Change the wording
Important
forcats::fct_recode will leave the levels that aren’t explicitly mentioned as is, and will warn you if you accidentally refer to a level that doesn’t exist.
To combine groups, you can assign multiple old levels to the same new level, or… use forcats::fct_collapse!
Dates and Time
In the Tidyverse, the main package to manage dates and time is lubridate.
Remind
- Dates are counts (based on ?doubles) of days since 1970-01-01.
- Date-Time are counts (based on ?doubles) of seconds since 1970-01-01.
To get the current date or date-time you can use today() or now():
Dates and Time - conversion from strings
Dates
[1] "2017-01-31"
[1] "2017-01-31"
[1] "2017-01-31"Dates-time
[1] "2017-01-31 20:11:59 UTC"
[1] "2017-01-31 20:11:00 UTC"
[1] "2017-01-31 20:00:00 UTC"
[1] "2017-01-31 UTC"Date <-> Date-time conversion
Extracting/Changing components
We can extract or modify components from date/date-time objects using:
year()month()day()hour()minute()second()-
wday()(day of the week) -
yday()(day of the year) -
week()(week of the year) -
quarter()(quarter of the year).
Extract
[1] "2024-02-29 11:16:17 CET"
[1] 2024
[1] 2
[1] 29
[1] 11
[1] 16
[1] 17.83648
[1] 5
[1] 60
[1] 9
[1] 1Change
[1] "2020-02-29 11:16:17 CET"
[1] "2020-12-29 11:16:17 CET"
[1] "2020-12-30 11:16:17 CET"
[1] "2020-12-30 17:16:17 CET"
[1] "2020-12-30 17:14:17 CET"
[1] "2020-12-30 17:14:56 CET"Your turn (main: B; bk1: C; bk2: A)
Your turn
Connect to our pad (https://bit.ly/ubep-rws-pad-ed3)
Connect to the Day-4 project in RStudio cloud (https://bit.ly/ubep-rws-rstudio)
…and:
Under the sections
4.2. Ex26 Ex27of the pad, write (in a new line) your answer to the questions reported.Then, open the scripts
19-factors.Rand20-date-time.Rand follow the instruction step by step.
15:00
Tip
factorsfrom base R, orforcats::fctfrom forcats are the best way to represent categories in R. They work similarly, but forcats is more informative and more flexible.DateandDate-timeare counts of days/seconds since 1970-01-01. Managing them in R is not easy, but lubridate makes it easier.
My turn
YOU: Connect to our pad (https://bit.ly/ubep-rws-pad-ed3) and write there questions & doubts (and if I am too slow or too fast)
ME: Connect to the Day-4 project in RStudio cloud (https://bit.ly/ubep-rws-rstudio): script 14-factors.R
Strings - Regular Expressions
Regular expressions are a powerful tool for matching text patterns. They are used in many programming languages to find and manipulate strings, and in R are implemented in the stringr package.
Base syntax for regular expressions
-
.matches any character -
*matches zero or more times -
+matches one or more times -
?matches zero or one time -
^matches the start of a string -
$matches the end of a string -
[]matches any one of the characters inside -
[^]matches any character not inside the square brackets -
|matches the pattern either on the left or the right -
()groups together the pattern on the left and the right
Example
The following match any string that:
-
acontainsa(str_view("banana", "a"): ) -
^astarts witha -
a$ends witha -
^a$starts and ends witha -
^a.*a$starts and ends witha, with any number of characters in between -
^a.+a$starts and ends witha, with at least one character in between -
^a[bc]+a$starts and ends witha, with at least oneborcin between -
^a(b|c)d$starts witha, followed by eitherborc, followed by an endingd.
Tip
To match special characters, you need to escape them with a double backslash (\\). I.e., you need to use \\., \\*, \\+, \\?, \\^, \\$, \\[, \\], \\|, \\(, \\).
To match a backslash, you need \\\\.
Strings - {stringr}
The stringr package provides a consistent set of functions for working with strings, and it is designed to work consistently with the pipe.
Functions
-
str_detect(): does a string contain a pattern? -
str_which(): which strings match a pattern? -
str_subset(): subset of strings that match a pattern -
str_sub(): extract a sub-string by position -
str_replace(): replace the first match with a replacement -
str_replace_all(): replace all matches with a replacement -
str_remove(): remove the first match -
str_remove_all(): remove all matches -
str_split(): split up a string into pieces -
str_extract(): extract the first match -
str_extract_all(): extract all matches -
str_locate(): locate the first match -
str_locate_all(): locate all matches -
str_count(): count the number of matches -
str_length(): the number of characters in a string
Tip
Because all stringr functions start with str_, in RStudio you can type str_ and then pressing TAB to see all its available functions.
Examples
Strings - concatenate
str_c: takes any number of vectors as arguments and returns a character vector of the concatenated values.str_glue: takes a string and interpolates values into it.
library(tidyverse)
tibble(
x = c("apple", "banana", "pear"),
y = c("red", "yellow", "green"),
z = c("round", "long", "round")
) |>
mutate(
fruit = str_c(x, y, z),
fruit_space = str_c(x, y, z, sep = " "),
fruit_comma = str_c(x, y, z, sep = ", "),
fruit_glue = str_glue("I like {x}, {y} and {z} fruits")
)Your turn (main: C; bk1: A; bk2: B)
Your turn
Connect to our pad(https://bit.ly/ubep-rws-pad-ed3)
Connect to the Day-4 project in RStudio cloud (https://bit.ly/ubep-rws-rstudio)
…and:
Before to evaluate it, in the pad, under the section
4.2. Ex8, write (in a new line) how can you match all files names that are R scripts (i.e., ending with.ror.R)? Report you option for a regular expression.Then, open the script
21-strings.Rand follow the instruction step by step.
10:00
Tip
All functions in stringr start with
str_, so you can typestr_and then pressingTABto see all its available functions.You can use
str_viewto see how a regular expression matches a string.str_glueis a powerful tool to concatenate strings and variables.
My turn
YOU: Connect to our pad (https://bit.ly/ubep-rws-pad-ed3) and write there questions & doubts (and if I am too slow or too fast)
ME: Connect to the Day-4 project in RStudio cloud (https://bit.ly/ubep-rws-rstudio): script 16-strings.R
Homework
Posit’s RStudio Cloud Workspace
- Project: Day-4
- Instructions:
- Go to: https://bit.ly/ubep-rws-website
- The text is the Day-4 assessment under the tab “Summative Assessments”.
- Script to complete on RStudio:
solution.R
Acknowledgment
To create the current lesson, we explored, used, and adapted content from the following resources:
The slides are made using Posit’s Quarto open-source scientific and technical publishing system powered in R by Yihui Xie’s Kintr.
Additional resources
License
This work by Corrado Lanera, Ileana Baldi, and Dario Gregori is licensed under CC BY 4.0
References
Break
10:00

UBEP’s R training for supervisors